15MA033 Software: Negative Information Storage Model for Genomic Data
novel information storage model for genomic data that stores only differences and missing data was developed. This model also has the low physical storage and computational resource requirements of current compression systems, while improving the precision of the stored data. High fidelity data is critical for research, diagnosis, and therapy selection involved in precision medicine approaches to human diseases. Compared to the full storage model, data loading was much faster requiring hours instead of days, and the query times were similar or better, and the data base size was about 100 fold smaller.
COMMERCIAL OPPORTUNITY
- An estimated 228,000 genomes had been completely sequenced worldwide by 2014, and this number is expected to double every 12 months. Storage of genomic data can require up to 6 billion pieces of information per individual and 300 GB of data. Prices for storage currently range from $250-1000 per year for 1 TB of data (~3 to 4 individuals). This market is rapidly expanding, with numerous companies providing storage sytems for genomic DNA including Amazon Redshift, Google Genomics, Illumina BaseSpace, DNAnexus, and PierianDx.
- Current data storage products focus on storing only the ~0.1% of data that accounts for individual genetic variation, allowing high compression of data. Our novel storage model comprising both variant and missing data prevents severe loss of precision and with similar reduced storage and bandwidth requirements of current positive data storage systems.
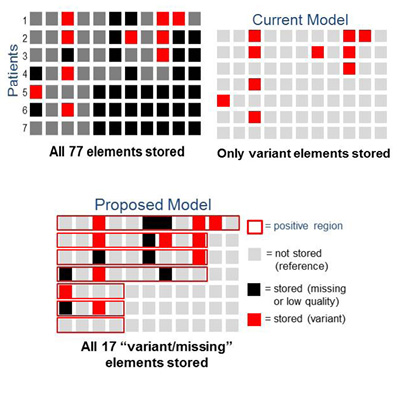
- In the example below, there is a small increase in physical storage demand (10 values stored in the Current Model B, compared with 17 values stored in the Proposed Model C) with a much greater precision for those data. These precise data will result in a greater quality of genomic information used as molecular markers for human diseases, such as cancer.
TECHNOLOGY
The model identifies a positive region (outlined in red ) of known data for each sample. The data within this region is compared to known reference values and is stored as “variant” only if it is different ( ) or as “missing” if it is absent or low quality ( ). No data are stored for unknown positions outside of this region that were never assayed ( ) or for data equivalent to the reference values within the positive region ( ). Also, contiguous data can be stored as regions to further compress the data within one patient subset. The storage model can subsequently be queried to determine any data value within the positive region.
PUBLICATION/PATENT
- U.S. Patent Application filed on 2/12/2018 for Dr. Jamie Teer
The Innovation Office
813.745.6828
InnovationMarketing@Moffitt.org

